First Report of Genetic Variability of Erysipelothrix sp. Strain 2 in Turkeys Associated to Vero Cells Morphometric Alteration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Identification of Erysipelothrix spp.
2.3. Transmission Electronic Microscopy (TEM)
2.4. Pulsed Field Gel Electrophoresis (PFGE)
2.5. Epidemiological and Statistical Analysis
2.6. Cellular Morphometry
2.7. Apoptosis
2.8. Invasion Test
2.9. Statistical Analysis
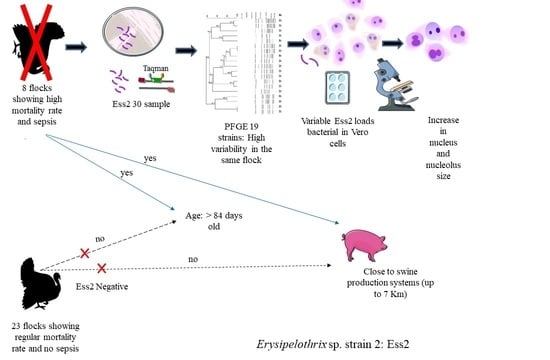
3. Results
3.1. Electron Microscopy of ES2
3.2. Epidemiologic Analysis Relative to PFGE
3.3. Morphometry in Vero Cells
3.4. Apoptosis
3.5. Bacterial Multiplication
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bricker, J.M.; Saif, J.M. Erysipelas. In Diseases of Poultry, 13th ed.; Swayne, D.E., Ed.; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Hollifield, J.L.; Cooper, G.L.; Charlton, B.R. An outbreak of erysipelas in 2-day-old poults. Avian Dis. 2000, 44, 721. [Google Scholar] [CrossRef] [PubMed]
- To, H.; Sato, H.; Tazumi, A.; Tsutsumi, N.; Nagai, S.; Iwata, A.; Nagano, T. Characterization of Erysipelothrix rhusiopathiae strains isolated from recent swine erysipelas outbreaks in Japan. J. Vet. Med. Sci. 2012, 74, 949–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feasi, M.; Pontali, E.; Usiglio, D.; Mori, M.; Cassola, G. Erysipelothrix rhusiopathiae septicaemia in systemic lupus erythematosus. Infez. Med. 2018, 26, 356–358. [Google Scholar] [PubMed]
- Kobayashi, K.; Kawano, T.; Mizuno, S.; Kubo, K.; Komiya, N.; Otsu, S. Erysipelothrix rhusiopathiae bacteremia following a cat bite. IDCases 2019, 18, e00631. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.E.; Richards, J.S.; Kerr, G.S.; Kan, V.L. Erysipelothrix rhusiopathiae septic arthritis. Arthritis Rheum. 2003, 48, 1156–1157. [Google Scholar] [CrossRef]
- Romney, M.; Cheung, S.; Montessori, V. Erysipelothrix rhusiopathiae endocarditis and presumed osteomyelitis. Can. J. Infect. Dis. 2001, 12, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Khan, D.; Mobarakai, N. Erysipelothrix rhusiopathiae endocarditis. IDCases 2020, 22, e00958. [Google Scholar] [CrossRef]
- Brooke, C.J.; Riley, T.V. Erysipelothrix rhusiopathiae: Bacteriology, epidemiology and clinical manifestations of an occupational pathogen. J. Med. Microbiol. 1999, 48, 789–799. [Google Scholar] [CrossRef] [Green Version]
- Verbarg, S.; Rheims, H.; Emus, S.; Frühling, A.; Kroppenstedt, R.M.; Stackebrandt, E.; Schumann, P. Erysipelothrix inopinata sp. nov., isolated in the course of sterile filtration of vegetable peptone broth, and description of Erysipelotrichaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 221–225. [Google Scholar] [CrossRef]
- Takahashi, T.; Fujisawa, T.; Tamura, Y.; Suzuki, S.; Muramatsu, M.; Sawada, T.; Benno, Y.; Mitsuoka, T. DNA Relatedness among Erysipelothrix rhusiopathiae strains representing all twenty-three serovars and Erysipelothrix tonsillarum. Int. J. Syst. Bacteriol. 1992, 42, 469–473. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Fujisawa, T.; Umeno, A.; Kozasa, T.; Yamamoto, K.; Sawada, T. A taxonomic study on erysipelothrix by DNA-DNA hybridization experiments with numerous strains isolated from extensive origins. Microbiol. Immunol. 2008, 52, 469–478. [Google Scholar] [CrossRef]
- Bang, B.-H.; Rhee, M.-S.; Chang, D.-H.; Park, D.-S.; Kim, B.-C. Erratum to: Erysipelothrix larvae sp. nov., isolated from the larval gut of the rhinoceros beetle, Trypoxylus dichotomus (Coleoptera: Scarabaeidae). Antonie Leeuwenhoek 2016, 109, 167–168. [Google Scholar] [CrossRef]
- Pomaranski, E.K.; Griffin, M.J.; Camus, A.C.; Armwood, A.R.; Shelley, J.; Waldbieser, G.C.; LaFrentz, B.R.; García, J.C.; Yanong, R.; Soto, E. Description of Erysipelothrix piscisicarius sp. nov., an emergent fish pathogen, and assessment of virulence using a tiger barb (Puntigrus tetrazona) infection model. Int. J. Syst. Evol. Microbiol. 2020, 70, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Janßen, T.; Voss, M.; Kühl, M.; Semmler, T.; Philipp, H.-C.; Ewers, C. A combinational approach of multilocus sequence typing and other molecular typing methods in unravelling the epidemiology of Erysipelothrix rhusiopathiae strains from poultry and mammals. Vet. Res. 2015, 46, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forde, T.; Biek, R.; Zadoks, R.; Workentine, M.L.; De Buck, J.; Kutz, S.; Opriessnig, T.; Trewby, H.; van der Meer, F.; Orsel, K. Genomic analysis of the multi-host pathogen Erysipelothrix rhusiopathiae reveals extensive recombination as well as the existence of three generalist clades with wide geographic distribution. BMC Genom. 2016, 17, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muellner, P.; Pleydell, E.; Pirie, R.; Baker, M.G.; Campbell, D.; Carter, P.E.; French, N.P. Molecular-based surveillance of campylobacteriosis in New Zealand—From source attribution to genomic epidemiology. Eurosurveillance 2013, 18, 1–7. [Google Scholar] [CrossRef]
- Hoepers, P.G.; dos Reis, T.F.M.; Mendonça, E.P.; Rossi, D.A.; Koerich, P.K.; França, T.V.J.; Zuffo, J.P.; Junior, E.C.V.; Fonseca, B.B. First outbreak reported caused by Erysipelothrix species strain 2 in turkeys from poultry-producing farms in Brazil. Ann. Microbiol. 2019, 69, 1211–1215. [Google Scholar] [CrossRef]
- Bozzola, J.; Russell, L. Specimen Preparation for Transmission Electron Microscopy. In Electron Microscopy; Sudbury, J.B., Ed.; CRC Press: Boca Raton, FL, USA, 1999; pp. 49–72. [Google Scholar]
- Centers for Disease Control and Prevention. Standardized Laboratory Protocol for Molecular Subtyping of Campylobacter Jejuni by Pulsed Field Gel Electrophoresis (PFGE). In PulseNet: The National Molecular Subtyping Network for Foodborne Disease Survellance; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013. [Google Scholar]
- Boerner, L.; Nevis, K.R.; Hinckley, L.S.; Weber, E.S.; Frasca, S. Erysipelothrix Septicemia in a Little Blue Penguin (Eudyptula Minor). J. Vet. Diagn. Investig. 2004, 16, 145–149. [Google Scholar] [CrossRef] [Green Version]
- Grazziotin, A.L.; Vidal, N.M.; Hoepers, P.G.; Reis, T.F.M.; Mesa, D.; Caron, L.F.; Ingberman, M.; Beirão, B.C.B.; Zuffo, J.P.; Fonseca, B.B. Comparative genomics of a novel clade shed light on the evolution of the genus Erysipe-lothrix and characterise an emerging species. Sci. Rep. 2021, in press. [Google Scholar]
- Bender, J.S.; Shen, H.G.; Irwin, C.K.; Schwartz, K.J.; Opriessnig, T. Characterization of Erysipelothrix species isolates from clinically affected pigs, environmental samples, and vaccine strains from six recent swine erysipelas outbreaks in the united states. Clin. Vaccine Immunol. 2010, 17, 1605–1611. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, H.; Bagge, E.; Båverud, V.; Fellström, C.; Jansson, D.S. Erysipelothrix rhusiopathiae contamination in the poultry house environment during erysipelas outbreaks in organic laying hen flocks. Avian Pathol. 2014, 43, 231–237. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Chang, B.J.; Riley, T.V. Erysipelothrix rhusiopathiae. Vet. Microbiol. 2010, 140, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Cross, G.M.J.; Claxton, P.D. Serological classification of australian strains of Erysipelothrix rhusiopathiae isolated from pigs, sheep, turkeys and man. Aust. Vet. J. 1979, 55, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Menzies, B.E.; Kourteva, I. Staphylococcus aureus α-toxin induces apoptosis in endothelial cells. FEMS Immunol. Med. Microbiol. 2000, 29, 39–45. [Google Scholar] [CrossRef]
- Tsai, P.-J.; Lin, Y.-S.; Kuo, C.-F.; Lei, H.-Y.; Wu, J.-J. Group a streptococcus induces apoptosis in human epithelial cells. Infect. Immun. 1999, 67, 4334–4339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, Y.; Ooka, T.; Shi, F.; Ogura, Y.; Nakayama, K.; Hayashi, T.; Shimoji, Y. The genome of Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, reveals new insights into the evolution of firmicutes and the organism’s intracellular adaptations. J. Bacteriol. 2011, 193, 2959–2971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, J.; Zhang, A.; Zhu, W.; Zhang, Q.; Xu, Z.; Yan, S.; Sun, X.; Chen, H.; Jin, M. iTRAQ-based quantitative proteomic analysis reveals potential virulence factors of Erysipelothrix rhusiopathiae. J. Proteomics 2017, 160, 28–37. [Google Scholar] [CrossRef]
- Zhu, W.; Cai, C.; Huang, J.; Liu, L.; Xu, Z.; Sun, X.; Jin, M. Characterization of pathogenic roles of two Erysipelothrix rhusiopathiae surface proteins. Microb. Pathog. 2018, 114, 166–168. [Google Scholar] [CrossRef]
- Fujisawa, S.; Romin, Y.; Barlas, A.; Petrovic, L.M.; Turkekul, M.; Fan, N.; Xu, K.; Garcia, A.R.; Monette, S.; Klimstra, D.S.; et al. Evaluation of YO-PRO-1 as an early marker of apoptosis following radiofrequency ablation of colon cancer liver metastases. Cytotechnology 2014, 66, 259–273. [Google Scholar] [CrossRef] [Green Version]
- Shimoji, Y.; Yokomizo, Y.; Mori, Y. Intracellular survival and replication of Erysipelothrix rhusiopathiae within murine macrophages: Failure of induction of the oxidative burst of macrophages. Infect. Immun. 1996, 64, 1789–1793. [Google Scholar] [CrossRef] [Green Version]
- Gallucci, S.; Lolkema, M.; Matzinger, P. Natural adjuvants: Endogenous activators of dendritic cells. Nat. Med. 1999, 5, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Rock, K.L. How dying cells alert the immune system to danger. Nat. Rev. Immunol. 2008, 8, 279–289. [Google Scholar] [CrossRef]
- Cullen, S.P.; Henry, C.M.; Kearney, C.J.; Logue, S.E.; Feoktistova, M.; Tynan, G.A.; Lavelle, E.C.; Leverkus, M.; Martin, S.J. Fas/CD95-induced chemokines can serve as “find-me” signals for apoptotic Cells. Mol. Cell 2013, 49, 1034–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimoji, Y. Pathogenicity of Erysipelothrix rhusiopathiae: Virulence factors and protective immunity. Microbes Infect. 2000, 2, 965–972. [Google Scholar] [CrossRef]
- Galán, J.E.; Cossart, P. Host–pathogen interactions: A diversity of themes, a variety of molecular machines. Curr. Opin. Microbiol. 2005, 8, 1–3. [Google Scholar] [CrossRef]
- Mostowy, S.; Cossart, P. Bacterial autophagy: Restriction or promotion of bacterial replication? Trends Cell Biol. 2012, 22, 283–291. [Google Scholar] [CrossRef]
- Díaz-Delgado, J.; Arbelo, M.; Sierra, E.; Vela, A.; Domínguez, M.; Paz, Y.; Andrada, M.; Domínguez, L.; Fernández, A. Fatal Erysipelothrix rhusiopathiae septicemia in two Atlantic dolphins (Stenella frontalis and Tursiops truncatus). Dis. Aquat. Organ. 2015, 116, 75–81. [Google Scholar] [CrossRef]






| Est06 | Est07 | LB | |
|---|---|---|---|
| Initial inoculum | 3.00 a | 3.00 a | 3.00 |
| Inside the cell culture medium | 7.22 b A | 7.11 b A | 0 |
| After four washes | 2.89 a B | 2.81 a B | 0 |
| After treatment with triton | 3.13 a C | 3.23 a C | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Reis, T.F.M.; Hoepers, P.G.; Peres, P.A.B.M.; Mendonça, E.P.; de Sousa Braga, P.F.; Beletti, M.E.; Rossi, D.A.; Grazziotin, A.L.; Goulart, L.R.; Fonseca, B.B. First Report of Genetic Variability of Erysipelothrix sp. Strain 2 in Turkeys Associated to Vero Cells Morphometric Alteration. Pathogens 2021, 10, 141. https://doi.org/10.3390/pathogens10020141
dos Reis TFM, Hoepers PG, Peres PABM, Mendonça EP, de Sousa Braga PF, Beletti ME, Rossi DA, Grazziotin AL, Goulart LR, Fonseca BB. First Report of Genetic Variability of Erysipelothrix sp. Strain 2 in Turkeys Associated to Vero Cells Morphometric Alteration. Pathogens. 2021; 10(2):141. https://doi.org/10.3390/pathogens10020141
Chicago/Turabian Styledos Reis, Thais Fernanda Martins, Patrícia Giovana Hoepers, Phelipe Augusto Borba Martins Peres, Eliane Pereira Mendonça, Paula Fernanda de Sousa Braga, Marcelo Emilio Beletti, Daise Aparecida Rossi, Ana Laura Grazziotin, Luiz Ricardo Goulart, and Belchiolina Beatriz Fonseca. 2021. "First Report of Genetic Variability of Erysipelothrix sp. Strain 2 in Turkeys Associated to Vero Cells Morphometric Alteration" Pathogens 10, no. 2: 141. https://doi.org/10.3390/pathogens10020141