The last universal common ancestor (LUCA) departed long ago, taking its evolutionary secrets to the grave or—more likely—the environs of a deep sea vent. Yet clues remain, scattered among the genomes of extant organisms. Such clues are often put together, figuratively speaking, by scientists who compare the genomes of organisms that split from the LUCA. But now select genomic clues have been put together in a way that could not be more literal. Specifically, genetic bequests long split between two domains of life, Bacteria and Archaea, have been reunited in a newly created organism.
Scientists from the University of Groningen and Wageningen University have engineered Escherichia coli so that it is more of an archaebacterium. They did so to test the hypothesis that Bacteria and Archaea evolved their separate ways because LUCA had an unstable cell membrane—an unstable mixture of lipids. It has long been assumed that intrinsic instability of mixed membranes led to a “lipid divide” and the subsequent differentiation of bacteria and archaea.
The scientists found that they could engineer cells that stably maintained a mixed heterochiral membrane. Essentially, the scientists refuted the old hypothesis. They also generated results that could advance membrane engineering in industrial settings, leading to the production of organisms with improved robustness.
Details of the scientists’ work appeared March 19 in the Proceedings of the National Academy of Sciences, in an article entitled “Converting Escherichia coli into an Archaebacterium with a Hybrid Heterochiral Membrane.” This article describes how the scientists constructed a stable hybrid heterochiral membrane through lipid engineering.
There are many ideas on how cellular life could have evolved billions of years ago. Protocells may have formed in clay minerals, or as simple lipid vesicles. In the latter scenario, something called the lipid divide would have occurred, creating the separate domains of Bacteria and Archaea, said Arnold Driessen, Ph.D., one of the authors of the current study and a professor of molecular microbiology at the University of Groningen: “The lipid membranes of both domains are different, composed of phospholipids that are each other's mirror image.”
In technical terms, the lipids in the membrane of bacteria are made up of straight-chain fatty acids that are ester-linked to a backbone of glycerol-3-phosphate. But the lipids in the archaeal membrane have a backbone of glycerol-1-phosphate, to which isoprenoids are linked by ether bonds.
The idea behind the lipid divide is that a common ancestor of both bacteria and archaea had a cell membrane in which both types of lipid were mixed. “This mixed membrane would be less stable than a homogeneous membrane of just one type of phospholipid, so eventually a split occurred, resulting in the two domains of Bacteria and Archaea,” explained Driessen.
All this would have happened over 3.5 billion years ago, so we can't expect any hard evidence to be around. Driessen and his co-workers therefore decided to reverse-engineer a microorganism with a mixed membrane. “This had been tried before,” he noted, “but these experiments resulted in bacteria with only very small amounts (<1%) of archaeal lipids.”
In the new study, the Groningen/Wageningen team brought this figure up to 30%.
“By boosting isoprenoid biosynthesis and heterologous expression of archaeal ether lipid biosynthesis genes, we obtained a viable E. coli strain of which the membranes contain archaeal lipids with the expected stereochemistry,” the PNAS article reported. “It has been found that the archaeal lipid biosynthesis enzymes are relatively promiscuous with respect to their glycerol phosphate backbone and that E. coli has the unexpected potential to generate glycerol-1-phosphate.”
These results were made possible by two key breakthroughs. First, the Groningen scientists discovered an enzyme that is crucial to the production of archaeal membrane lipids. “This takes three steps,” noted Driessen, “and before that only two of the enzymes involved were known.” Second, Wageningen scientists managed to increase the production of isoprenoids in the bacterium E. coli. Isoprenoids are a ubiquitous class of organic compounds that includes many natural flavors, colors, and fragrances.
Both discoveries were transferred to a normal E. coli bacterium, which was a major feat of genetic engineering. “And we didn't know if the end result would be a viable cell,” said Driessen. But in the end, after some fine tuning, a cell was created in which all phosphatidylglycerol, the lipids which form the basic bilayer of the bacterial membrane, was replaced by the archaeal equivalent (archaetidylglycerol).
“This bacterium grew at normal speed and was stable,” declared Driessen. “So this result does not support the hypothesis that a mixed membrane is inherently instable and could thus have created the lipid divide.”
Driessen noted that the archaeal enzymes for membrane lipid production are less specific in the reactions they catalyze than their bacterial equivalents. (They are described as being more “primordial” in the PNAS article.) So the evolution of enzyme specificity could have been a driver for the divide. There is of course one major caveat. The experiments were done in modern E. coli bacteria, which have evolved 3.5 billion years beyond the original split with the archaea.
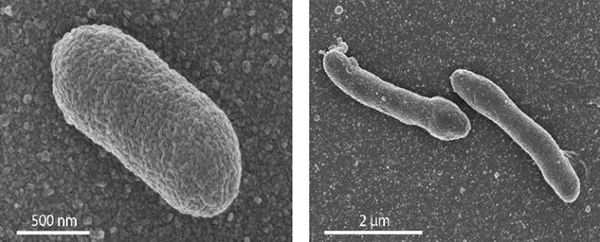
“The robustness of these mixed cells surprised us,” Driessen admitted. “We expected more problems keeping them alive. After all, what we have engineered does amount to a new life form.” The engineered cells were more elongated than the original E. coli. And when the production of archaeal lipids was very high, growth slowed and the membrane developed lobular appendages.
Apart from the evolutionary implications, this discovery, Driessen suggested, could spawn new research: “For example, we could engineer a bacterial expression system for archaeal membrane proteins, such as those produced by hyperthermophiles that grow at extremely high temperatures and pressure.”